The Datasets
Living datasets backed by automated experiments and open source methods.
DATASET
Protein Sequence to Expression
Protein expression is a fundamental process in biotechnology- crucial for academic research, human health applications, and the bioeconomy. However, predicting which proteins will express well in different organisms remains a significant challenge. Current approaches often rely on laborious experimental trial-and-error methods.
A generalizable model for predicting protein expression from sequence information would revolutionize fields ranging from basic scientific research to biomanufacturing and pharmaceutical development.
READ
Can protein expression be “solved”? (A review of the current state of protein expression data)
A strategy for scalable data collection of soluble protein expression in diverse hosts (Our proposed experimental approach)
We propose to develop a large-scale, standardized dataset enabling quantitative prediction of expression levels from protein sequence data across multiple organisms. This dataset will serve as a foundation for machine learning models that can accurately predict soluble protein expression, addressing a major bottleneck in biotechnology and protein engineering.
OUR STRATEGY
Protein expression is the result of the complex interplay of intrinsic and extrinsic factors. Our strategy is to create a comprehensive dataset linking protein sequences to expression levels in various organisms to enable the development of accurate, generalizable predictive models for protein expression.
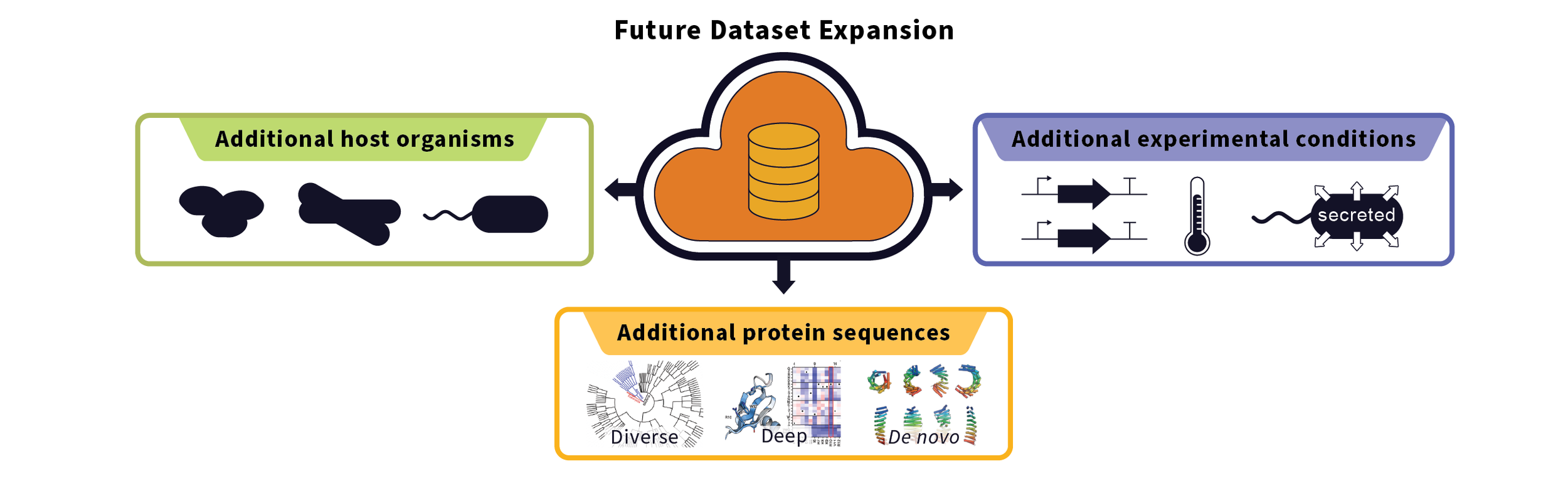
Initially, we will focus on expression data in E. coli and P. pastoris, two widely used expression hosts. As the dataset grows, we will expand to include more diverse organisms, more sequences, and more experimental conditions to capture a broader range of expression behaviors.
HOW ARE WE DOING IT?
We are using a combination of high-throughput pooled methods and targeted validation to collect expression data on over 1 million unique protein open reading frames. Our approach includes:
Pooled methods: Label-free proteomics and SortSeq
Validation methods: HiBiT and mass spectrometry
This strategy allows us to generate a large, diverse dataset while ensuring data quality and reproducibility. Data will be freely available via API access, provided in a standardized format, and designed with ML utility in mind to promote widespread use and collaboration.
OUR TIMELINE
Initial timelines for the protein expression dataset project:
2024: Establish experimental protocols and data collection pipeline in E. coli
2025: Expand data collection to P. pastoris and begin large-scale data generation
2026+: Introduce additional validation methods and start expanding to additional bacterial and eukaryotic hosts
Project Team
Kasia Baranowski
TECHNICAL PROJECT MANAGER
Team Member Spotlight
Kasia attended graduate school at Harvard where she studied the mycobacterial cell wall in the lab of Eric Rubin. Since then, she’s worked in synthetic biology as a yeast strain engineer (Zymergen), a CRISPR platform biologist (Inscripta) and a scientific project manager (Cemvita). At Align to Innovate, Kasia spearheads the Protein Sequence to Expression dataset. She wants you to stop wasting time trying to express proteins in the wrong host and knows the expression dataset and subsequent models will help you out!
Proposal Contributors
Eli Bixby - Cradle Bio
Swati Choudhary - Formerly at Shiru, Inc.
Ranjani Varadan - Formerly at Shiru, Inc.
Elise de Reus - Cradle Bio
Aljaž Gaber - University of Ljubljana, Faculty of Chemistry and Chemical Technology
Sebastian Jaaks-Kraatz - Friedrich Miescher Institute for Biomedical Research
Michael C. Jewett - Department of Bioengineering, Stanford University
Ben Lehner - Center for Genomic Regulation
Hector Garcia Martin - Lawrence Berkeley National Laboratory, DOE Joint BioEnergy Institute, DOE Agile BioFoundry
Evangelos-Marios Nikolados - School of Biological Sciences, University of Edinburgh
Diego A. Oyarzún - School of Informatics & School of Biological Sciences, University of Edinburgh
Christopher J. Petzold - Lawrence Berkeley National Laboratory
Christopher R. Reynolds - Eden Genetics Ltd
David Ross - National Institute of Standards and Technology
Howard Salis - Pennsylvania State University
Devin Scannell - Independent
Rachel Sevey - Independent Graphic Design and Data Visualization Specialist
Data Collection Partners
Talk to us! Here’s how to participate:
Email us at datasets@alignbio.org