Bijoy is a biochemist turned synthetic biologist with a passion for advancing innovative ideas. His work spans industry and academia, from applying generative AI and NGS to develop vaccines, therapies, and diagnostics, to using genome engineering and single-molecule biophysics to study protein synthesis. As Technical Project Manager at the Align Foundation, he is leading the development and growth of Protein Engineering Tournaments as a model for collaborative benchmarking in protein design.
A transparent platform for fostering collaboration, evaluating progress, and establishing benchmarks.
💰 $30,000 in Cash Prizes 🧬 Free DNA Synthesis + Wet Lab Testing 📣 Publish Your Results
The Protein Engineering Tournament is a global benchmarking initiative hosted by The Align Foundation. This community-driven challenge empowers researchers to evaluate and improve predictive and generative models in protein engineering. Like CASP for protein structure prediction, the Tournament is designed to foster collaboration, transparency, and rapid progress in AI-powered protein design.
2025 Target
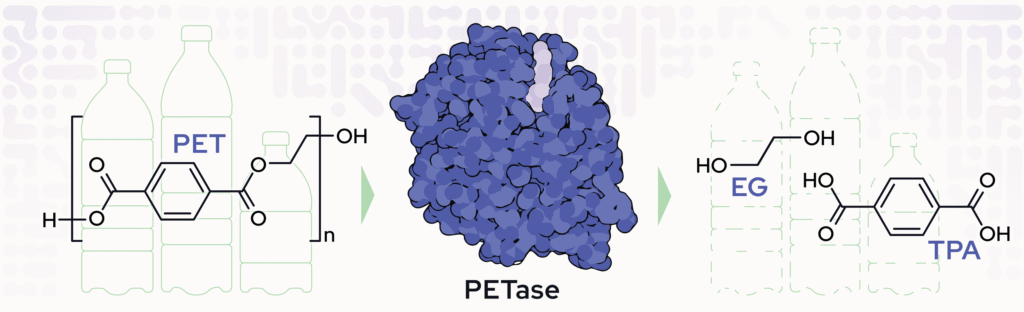
This year’s Tournament centers on polyethylene terephthalate hydrolase (PETase), an enzyme with real-world impact in the plastic waste crisis, and the circular plastic economy.
HOW IT WORKS
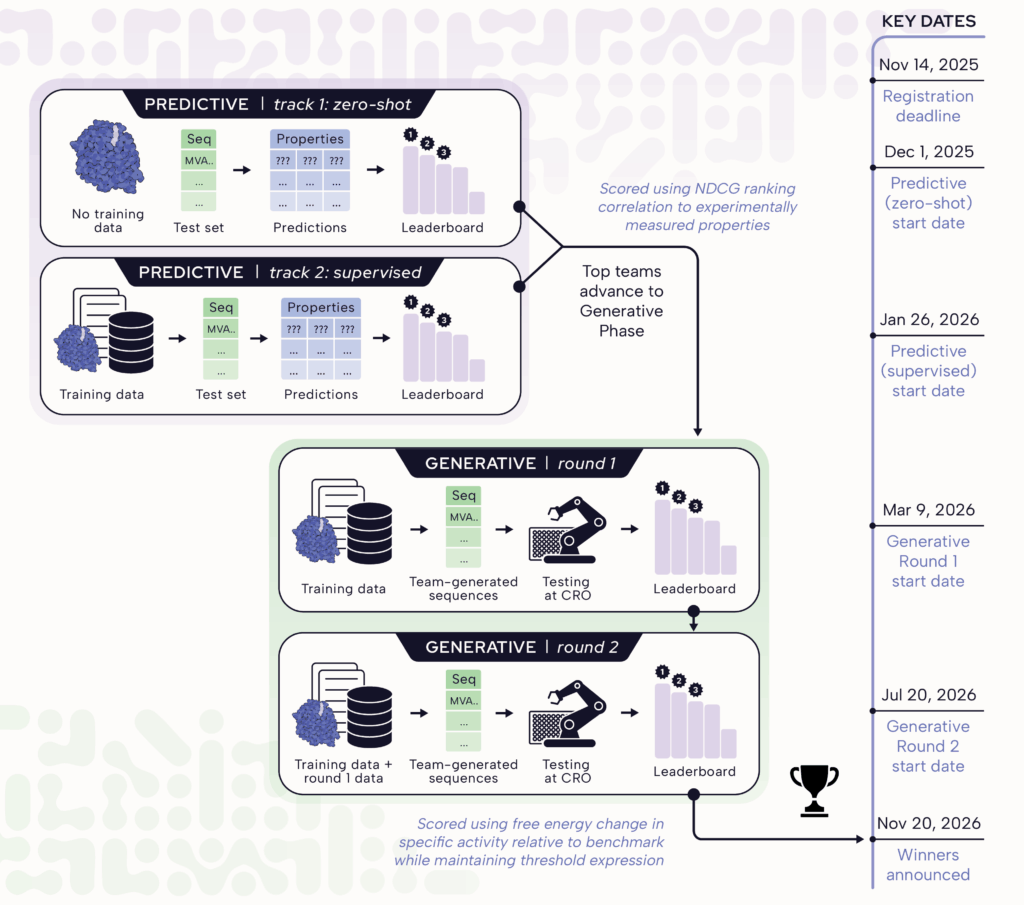
Tournament Phases & Timeline
Evaluation Criteria
- Predictive Phase: For both zero-shot, and supervised track teams will be ranked using Normalized Discounted Cumulative Gain (NDCG), which measures how well participant-submitted prediction rankings correlate with ground truth experimental rankings. The final tournament ranking is determined by the average NDCG across all protein properties. In the event of tied scores, rankings are determined by submission time, with earlier submissions taking precedence.
- Generative Phase: At the end of round-2 teams will be ranked by comparing free energy change of the experimentally measured specific activity relative to benchmark value, while maintaining threshold levels of expression.
– Overall winner: Free energy change of specific activity relative to benchmark value, averaged over the top 10% of sequences
– Best sequence: Sequence with highest free energy change of specific activity relative to benchmark value.

Incentives & Awards
- $30,000 in cash prizes
-
DNA synthesis and experimental testing of your designs
-
Leader-board recognition alongside leading researchers
- Access to high quality protein engineering datasets
-
Co-authorship in a special issue publication
-
Invitation to present at key conference
-
Impact: Help solve plastic waste crisis
Program Manager

Our Sponsors

Evolutionary Scale

Modal

NVIDIA Health

Twist Bioscience - Synthesis Sponsor
Frequently Asked Questions
No. Participation in the Tournament is free of charge.
Anyone. We welcome teams from academia, industry, nonprofits, and independent participants worldwide.
You can register solo. We’ll help connect you with others or you can form your own team later.
Due to limited capacity, only top performing teams from both zero-shot and supervised tracks of the Predictive Phase will be invited to participate in the Generative Phase. The best way to participate in the Generative Phase is to perform well in either of the Predictive Phase tracks.
- Predictive Phase (Zero-Shot Track): We will provide protein sequences for which we seek prediction of properties like activity, thermostability, or expression
- Predictive Phase (Supervised Track): We will provide protein sequences for which we seek prediction of properties like activity, thermostability, or expression. We will provide a training dataset of activity, thermostability, and expression of natural PETase sequences and mutational variants.
- Generative Phase (Round 1): We will provide a training dataset of activity, thermostability, and expression of natural PETase sequences and mutational variants.
- Generative Phase (Round 2): In addition to the training dataset, we will also provide results of experimentally measured activity, thermostability, and expression data of the designed protein sequences submitted by each team.
The Align Foundation will own those datasets, methods, assays, analysis and results (“Outputs”) generated for your submissions. This is to permit Align to freely organize and publish Outputs as that is the primary goal for the Tournament. Participants retain IP rights to all protein sequences they design, algorithms they develop, and novel methods they use to generate top line Outputs. There is a license-back mechanism to resolve any overlap between Outputs and IP Participants would like to own.
If you create a novel, industrially relevant PETase or powerful new method to design one during the Tournament you should patent it!
Read the details of our IP policy below.
Anonymized predictions, designed protein sequences, and associated experimental results for all teams will made public at the completion of the Tournament.
Top performing teams will be de-anonymized after the completion of the Tournament.
Teams will be anonymized during the Tournament. Top-performing teams will be de-anonymized at the end of the Tournament.
- Each team submits their property predictions (predictive phase) or designed protein sequences (generative phase). You also must submit an abstract describing your modeling approach.
- Sharing detailed methods and deposition of code on Github is strongly encouraged but not required.
Still have questions? Email us at tournament@alignbio.org.
The Tournament is organized for advancing research purposes and public knowledge regarding the useful engineering of proteins. All teams must comply with our participation terms and conditions. Please see the full agreement here.





